Research Article - Onkologia i Radioterapia ( 2023) Volume 17, Issue 7
Classification of Ultrasound Breast Cancer Image Using Tuning Up the Hyper-Parameter of Convolutional Neural Network
Satish Bansal1*, Rakesh S Jadon2 and Sanjay Kumar Gupta32Department of Computer Engineering, MITS, Gwalior, India
3SOS in Computer Science & Applications, Jiwaji University, Gwalior, India
Satish Bansal, Prestige Institute of Management & Research, Gwalior, India, Email: satish_bansal@rediffmail.com
Received: 02-Jun-2023, Manuscript No. OAR-23-101024; Accepted: 20-Jul-2023, Pre QC No. OAR-23-101024 (PQ); Editor assigned: 18-Jun-2023, Pre QC No. OAR-23-101024 (PQ); Reviewed: 05-Jul-2023, QC No. OAR-23-101024 (Q); Revised: 18-Jul-2023, Manuscript No. OAR-23-101024 (R); Published: 23-Jul-2023
Abstract
Breast cancer in women is a significant public health concern worldwide, with many cases going undiagnosed until the advanced stages. Early detection is crucial for proper treatment and improved outcomes. There are some pre-trained models used by authors for the detection of breast tumour, but these models require extensive computation power due to their many layers and parameters. To address this issue, it is required to proposed Convolutional Neural Network (CNN) model with fewer training parameters for classification of ultrasound images dataset to determine that a particular image is either benign or malignant. In this paper, CNN model is proposed with changes in some hyper parameters like the number of filters, filter size, batch normalization, learning rate, epoch, and batch size, to achieve better accuracy with less computational power. The proposed model was compared to other pre-trained models, including ResNet50, Efficient Net, and VGG16, using two databases (database A for training and validation data, and database B for testing data). Our proposed classifier outperformed than pretrained classifiers in terms of accuracy.
Keywords
ultrasound image dataset, CNN, deep learning, breast cancer, CAD system, pre-trained CNN model
Introduction
Breast tumor is a widespread disease that distresses women in both developing and developed countries. Over the past decade, breast cancer has rapidly increased worldwide. Approximately more than 500 men and 41,000 women have died from breast tumor recently to the American Cancer Society (ACS). Breast cancer cells fall into two categories: benign and malignant. A benign tumor is not a cancerous cell and not dangerous for breast structure. In cases of malignant tumour spread to other body part and very harmful to other organs.
Medical professionals use various medical modalities to analyse and detect breast cancer, including X-ray, sonography etc. However, analysing these images is taking more time and is challenging for doctors to achieve accuracy. Initially, X-ray was the primary modality used for image processing to analyse breast cancer. Currently, ultrasound modality is most effective in analysing breast tumours due to its non-invasive and radiationfree nature [1], helping detect breast cancer and decreasing biopsies in females [2].
Machine Learning (ML) is also used to analyse medical images and try to detect many medical diseases, but due to some limitations like feature extraction from the image dataset, Deep Learning (DL) plays a noteworthy role in medical imaging modalities to analyse medical images. DL falls under machine learning, improving the performance of computer-aided diagnostic systems for medical problems. Deep learning is the scientific study of detecting and categorizing different types of health diseases like brain tumour, diabetes, and different types of cancer.
Various authors have used existing deep learning models to detect breast cancer, such as VGG, ResNet, and Inception. Since each Convolutional Neural Network (CNN) model uses different layers and many parameters, which are a combination of convolutional, relu, pooling, softmax, and fully connected layers, these pre-trained models take more time due to detecting many different objects on a large dataset.
Early detection of breast cancerous cells in ultrasound images benefits the early analysis of an unhealthy individual, improving the survival rate. Many authors have built CNN models for the analysis and recognition of breast cancer, but they fail to provide optimum outcomes due to the lack of pre-processing on the dataset, small dataset size, and inadequate training for feature extraction. To address these limitations, an effective and efficient CNN model is required.
Deep learning models are used in different domains, and medical science is an important area for researchers to find gaps or opportunities for better research. The purpose of using deep learning networks, researchers is to classify the prediction of patient diseases using medical images. Many authors focus on the accuracy metrics, but it is not sufficient. The overall classification of any deep learning model depends on the confusion matrix, F score, and AUC curve as well.
This provides an opportunity for new researchers to develop an effective CNN model in different areas, with the main focus on improving accuracy without increasing the parameters of deep learning models for multi-classification.
Review of Literature
Authors used “Histopathological Image Analysis” (HIA) for breast cancer classification using different techniques of ANN and CNN [3]. In their study, the authors used CNN algorithm on eight datasets set using the cross-validation method [4]. AUC was used as an indicator of accuracy.
The fast-growing population has led to an exponential increase in medical images, and traditional methods are failed to detect the breast cancer with the increasing demands for medical images [5]. In Mahmood M, et al., authors demonstrated CNN model in the classification of cancer and tumours cells [6].
In Fatima N, et al., authors reviewed many research papers on supervised learning and deep neural network for predicting the breast tumour [7]. Many authors introduced CNN models to detect and classify breast tumors and increase diagnosis efficiency [8-10].
Researchers used different pre-trained CNN models with fine tunes for different classifications [11-13]. They found training speed and accuracy can be improved through these pre-trained models with transfer learning.
Many authors developed the architecture of automatic prediction of normal, benign and malignant breast tumour [14]. In the past, researchers use advanced techniques for analysing breast cancer cells and classified them into benign and malignant [15-17].
The different methods and accuracy used by different authors on image dataset shown in Table 1.
Tab. 1. Previous study on breast cancer detection by different authors
| Reference | Year | Methods | Accuracy | Image Dataset |
|---|---|---|---|---|
| [18] | 2020 | U-Net | 98.59% | Ultrasound Image UI) |
| [19] | 2018 | Alex-Net | 91% | (UI) |
| [20] | 2021 | CNN and Deep representation Scaling | 91.5% | (UI) |
| [21] | 2016 | Optimized Feedforward Artificial Neural Network | 89.77% | Magnetic Resonance Imaging (MRI) |
| [22] | 2020 | Deep Neural Network with Support Value | 97.21% | Histopathology Image |
| [23] | 2021 | Inception V3 Res-Net 50 VGG 19 |
96% 94% 95% |
(UI) |
Objectives
In this paper, main aim to address the research gap between the creation of a proposed CNN model and a pre-trained CNN model. This paper opens several questions for researchers to explore:
1. Which is better for a specific problem creating a proposed CNN model with tuning up hyper-parameters or using a pretrained model with transfer learning?
2. How important is the number of layers compared to the number of parameters for the best classification in the CNN model?
3. Does a CNN model depend solely on accuracy or other performance measures? Does it also give promising results on testing data?
In order to develop a robust system that can give the best prediction on test dataset, we have the following objectives in this study:
1. Improving the accuracy of the classification and saving the computational power by the proposed classifier with a small number of layers and parameters. Compare with another pretrained CNN model.
2. This study used the dataset of breast cancer ultrasound images and tuned up the hyper parameters to make the efficient proposed model.
3. The classification measures of the proposed classifier and compare their results with another pre-trained CNN model.
Proposed Methodology
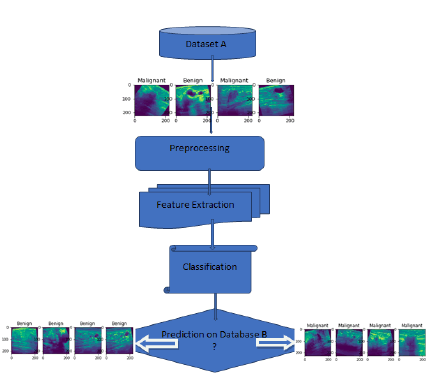
Figure 1 depicts the proposed workflow, which utilizes two breast ultrasound image datasets: database A and database B. The proposed CNN model incorporates common classification steps, including pre-processing, feature extraction, and classification. Various layer combinations, along with appropriate hyperparameters, are used to develop a robust model that effectively mitigates overfitting and bias in the dataset. By leveraging these techniques, the study aims to maintain the best classification accuracy while reducing computational requirements.
Figure 1: Flow of Proposed Work
Dataset
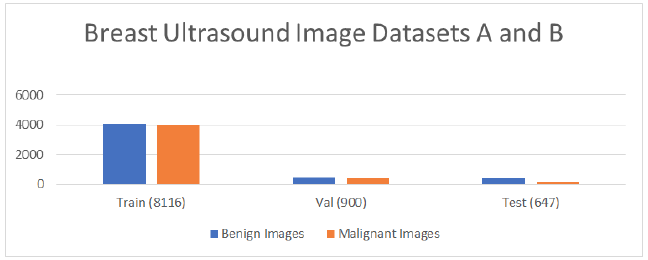
The proposed CNN model utilized two databases, A and B. For image analysis, the breast ultrasound image dataset A was divided into two directories, namely train and val [24]. Each directory contained two classes, namely benign and malignant, which contained real grayscale images. Dataset A consisted of 9016 images, as shown in Table 2. Dataset B was divided into three directories, namely benign, malignant, and normal [25]. Each directory contained two types of images, i.e., real grayscale and mask image. Select only two directories benign and malignant real images shown in Table 3, which were collected for testing the dataset by using the proposed classifier. In this paper, a proposed classifier is developed and applied to both types of datasets separately for classification, and the results were analysed.
Tab. 2. Ultrasound Image Dataset A
| Dataset A (9016) | Train (8116) | Val (900) |
|---|---|---|
| Benign Images (4574) | 4074 | 500 |
| Malignant Images (4442) | 4042 | 400 |
Tab. 3. Ultrasound Image Dataset B
| Dataset B | Test (647) |
|---|---|
| Benign Images | 437 |
| Malignant Images | 210 |
The graphical representation of datasets A and B is shown in Figure 2, which is divided into Benign, Malignant. Figure 3 shows the random sample of real grayscale ultrasound images from database A.
Figure 2: Datasets A and B.
Figure 3: Sample of Ultrasound Images
Convolutional Neural Network (CNN) model
The proposed CNN model uses different packages and libraries of Python 3.6. For the experiment, architecture of the projected classifier is depicted in Table 4.
Tab. 4. Architecture of Proposed CNN Model "sequential_1"
| Layer (type) | Output Shape | Param # |
|---|---|---|
| conv2d_4 (Conv2D) | (None, 224, 224, 32) | 320 |
| batch_normalization_4 | (Batch (None, 224, 224, 32) | 128 |
| max_pooling2d_4 (MaxPooling2) | (None, 112, 112, 32) | 0 |
| conv2d_5 (Conv2D) | (None, 112, 112, 64) | 18496 |
| batch_normalization_5 (Batch) | (None, 112, 112, 64) | 256 |
| max_pooling2d_5 (MaxPooling2) | (None, 56, 56, 64) | 0 |
| conv2d_6 (Conv2D) | (None, 56, 56, 128) | 73856 |
| batch_normalization_6 (Batch | (None, 56, 56, 128) | 512 |
| max_pooling2d_6 (MaxPooling2 | (None, 28, 28, 128) | 0 |
| dropout_2 (Dropout) | (None, 28, 28, 128) | 0 |
| conv2d_7 (Conv2D) | (None, 28, 28, 256) | 295168 |
| batch_normalization_7 (Batch | (None, 28, 28, 256) | 1024 |
| max_pooling2d_7 (MaxPooling2 | (None, 14, 14, 256) | 0 |
| flatten_1 (Flatten) | (None, 50176) | 0 |
| dropout_3 (Dropout) | (None, 50176) | 0 |
| dense_1 (Dense) | (None, 1) | 50177 |
Depending on the nature of the particular problem, the pretrained CNN architecture can be customized through transfer learning to create a revised pre-trained classifier. However, in this study, a proposed CNN model was created instead of using a pretrained model. The proposed CNN model consists of a lesser number of layers and parameters as shown in Table 5, which helps in saving computational power
Tab. 5. Comparison of CNN models based on Layers and Parameters
| CNN Model | Layers | Total Parameters | Trainable Parameters | Non-Trainable Parameters |
|---|---|---|---|---|
| Resnet50 | 50 Layers | 23,688,065 | 100,353 | 23,587,712 |
| VGG16 | 16 Layers | 14,739,777 | 25,089 | 14,714,688 |
| Efficient Net B7 | 813 Layers | 64,223,128 | 125,441 | 64,097,687 |
| New CNN Model | 16 Layers | 439,937 | 960 | 438,977 |
The proposed classifier uses a few hyper-parameters to improve the efficiency and reliability of the model. Table 6 highlights the following hyper-parameters used in the proposed CNN. The ideal approach in the experiment is to tune up hyperparameters and make some changes in the training options to build an effective CNN model. The training data use the following options for training: epochs are 30, learning_rate is 0.009 and the batch_size is 32.
Tab. 6. Tuning Up Hyper-Parameter in Proposed CNN
| Hyper-Parameter | Value | Purpose |
|---|---|---|
| Batch_Size | 32 | Control the number of training data |
| Learning_rate | 0.009 | Used to update the weights during training |
| Dropout layer | 0.2 | Control and preventing the overfitting |
| Batchnormalization layer | Speed up training and improve accuracy | |
| Pooling size | Max (2, 2) | To reduce the numbers of parameters/dimension |
| Kernel size | (3, 3) | It helps to generalize better |
Results
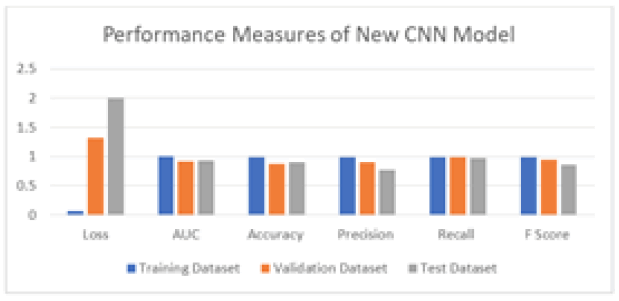
Table 7 displays the classification measures of the proposed classifier for the training, validation, and testing datasets, which are also represented in Figure 4. The proposed model automatically calculates the following matrices through model compilation during the training phase. The validation matrices are also calculated through the validation data. After learning from the training data, the model is evaluated on the testing dataset to determine its performance. The training, validation, and testing accuracies are 99%, 87%, and 89%, respectively. The proposed CNN model utilized 83% of the training data, 10% of the validation data from database A, and 7% of the testing data from database B, obtained from two different datasets.
Tab. 7. Performance Measure of proposed CNN Model for Epoch 30
| Performance Measures | loss | AUC | accuracy | precision | recall | F1 score |
|---|---|---|---|---|---|---|
| Training Data | 0.0699 | 0.9959 | 0.9901 | 0.9897 | 0.991 | 0.9903 |
| Validation Data | 1.3155 | 0.9129 | 0.8748 | 0.8975 | 0.99 | 0.9414 |
| Test Data | 2 | 0.9296 | 0.8949 | 0.7649 | 0.9762 | 0.8577 |
Figure 4: Performance Measures of New CNN Model for training, Validation and Test Dataset
Table 8 shows the loss and accuracy of ResNet50, VGG16 and EfficientNetB7 on the training and validation dataset at epoch 10. The proposed CNN model finds the best result as compared to these pre-trained models. The input size of the proposed classifier is 224 × 224 × 1 while the input size of the pre-trained classifier is 224 × 224 × 3. The accuracy of the proposed classifier is 99% which is high compared to other pre-trained models like Resnet50, VGG16 and EfficientNetB7.
Tab. 8. Comparison of pre-trained model with proposed CNN model at epoch 10.
| CNN Model Epoch (10) | Loss | accuracy | Validation loss | Validation accuracy |
|---|---|---|---|---|
| ResNet50 | 0.36 | 0.86 | 0.483 | 0.804 |
| VGG16 | 0.068 | 0.97 | 0.334 | 0.84 |
| EfficientNetB7 | 2.38 | 0.59 | 3.44 | 0.57 |
| Proposed CNN model | 0.093 | 0.9773 | 0.7942 | 0.8967 |
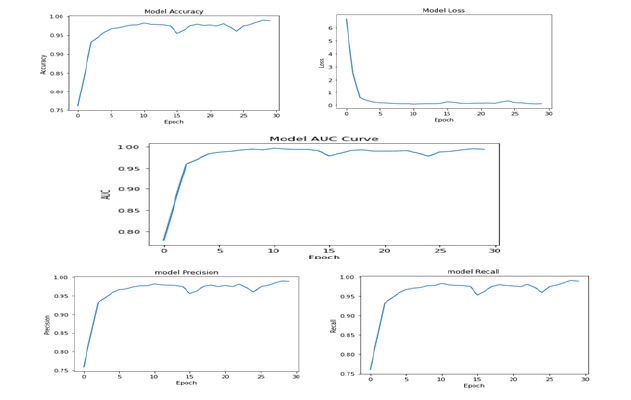
The training accuracy graph, loss graph, precision, recall graph and AUC graph are shown in Figure 5 at epoch 30. F1 score can be computed by using
Figure 5: Performance Metrics of Proposed CNN Model for the Epoch 30
F1 score = 2 × Precision × Recall / (Precision + Recall).
The performance measure of the CNN model is not solely dependent on accuracy, as other performance measures like F1 score and AUC curve are also important. The AUC, which represents area under the ROC curve, indicates how fit the model can predict on the testing data. The AUC values for the training dataset, validation dataset and testing datasets are 99%, 91%, and 92%, respectively, indicating that the model's performance is excellent. The F1 score considers false negatives and false positives and is a useful measure of the model's overall performance. The F1 score values for the training dataset, validation and testing datasets are 99%, 94%, and 85%, respectively, indicating that the model is giving promising results.
Discussion
There are many ways to create CNN models and the best model gives excellent results for the test data. The proposed model focused on fewer layers and parameters to save computational power. Tuning up the hyper-parameter in the proposed classifier maintains the maximum accuracy as shown in the result.
Conclusion
The proposed classifier uses a series of sequential steps to create an automated diagnosis system with modifications to hyperparameters. To ensure the robustness of the classifier, several experiments were performed to create a proposed deep learning model and also compared to other pre-trained CNN models. This scientific proof validates that the proposed CNN model for detecting breast cancer in ultrasound images provides greater accuracy after numerous experiments. In the future, a new framework could be developed to achieve two objectives, one is of perform semantic segmentation and classify. This could assist imaging specialists and doctors in detecting breast cancer in ultrasound images.
Declarations
Availability of supporting data
Support of data is from two references i.e, 24 and 25.
Competing Interest
No conf icts of interest.
Funding
No funding from any source.
Authors' Contributions
All authors contributed to the research methodology and analysis of results.
Acknowledgements
I’d like to thank my guides.
References
- Sun Q, Lin X, Zhao Y, Li L, Yan K, et al. Deep learning vs. radiomics for predicting axillary lymph node metastasis of breast cancer using ultrasound images: don't forget the peritumoral region. Front Oncol. 2020; 10:53.
- Cheng HD, Shan J, Ju W, Guo Y, Zhang L. Automated breast cancer detection and classification using ultrasound images: A survey. Pattern Recognit. 2010; 43:299-317.
- Zhou X, Li C, Rahaman MM, Yao Y, Ai S, et al. A comprehensive review for breast histopathology image analysis using classical and deep neural networks. IEEE Access. 2022; 8:90931-90956.
- Sutanto DH, Ghani MK. A benchmark of classification framework for non-communicable disease prediction: a review. ARPN J Eng Appl Sci. 2015; 10:9941-9955.
- Liu B, Yao K, Huang M, Zhang J, Li Y, et al. Gastric pathology image recognition based on deep residual networks. IEEE. 2018; 2: 408-412.
- Mahmood M, Al-Khateeb B, Alwash WM. A review on neural networks approach on classifying cancers. IAES Int J Artif Intell. 2020; 9:317.
- Fatima N, Liu L, Hong S, Ahmed H. Prediction of breast cancer, comparative review of machine learning techniques, and their analysis. IEEE Access. 2020; 8:150360-150376.
- Litjens G, Sánchez CI, Timofeeva N, Hermsen M, Nagtegaal I, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016; 6:26286.
- CireÅ?an DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Springer Berl Heidelb. 2013; 411-418.
- Chekkoury A, Khurd P, Ni J, Bahlmann C, Kamen A, et al. Automated malignancy detection in breast histopathological images. InMed. Imaging: Comput.-Aided Diagn. 2012; 8315: 332-344.
- Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. 2006; 18:1527-1554.
- Russakovsky O, Deng J, Su H, Krause J, Satheesh S, et al. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015; 115: 211-252.
- Oquab M, Bottou L, Laptev I, Sivic J. Learning and transferring mid-level image representations using convolutional neural networks. IEEE conference. 2014; 1717-1724.
- Huynh B, Drukker K, Giger MJ. MOâ?DEâ?207Bâ?06: Computerâ?aided diagnosis of breast ultrasound images using transfer learning from deep convolutional neural networks. Med Phys. 2016; 43:3705.
- George YM, Zayed HH, Roushdy MI, Elbagoury BM. Remote computer-aided breast cancer detection and diagnosis system based on cytological images. IEEE Syst J. 2013; 8:949-964.
- Jan Z, Khan SU, Islam N, Ansari MA, Baloch B. Automated detection of malignant cells based on structural analysis and naive Bayes classifier. Sindh Univ Res J. 2016; 48.
- McCann MT, Ozolek JA, Castro CA, Parvin B, Kovacevic J. Automated histology analysis: Opportunities for signal processing. IEEE Signal Process Mag. 2014; 32: 78-87.
- Hussain S, Xi X, Ullah I, Wu Y, Ren C, et al. Contextual level-set method for breast tumor segmentation. IEEE Access. 2020; 8:189343-189353.
- Yap MH, Pons G, Marti J, Ganau S, Sentis M, et al. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J Biomed Health Inform. 2017; 22:1218-1226.
- Byra M. Breast mass classification with transfer learning based on scaling of deep representations. Biomed Signal Process Control. 2021; 69:102828.
- Bevilacqua V, Brunetti A, Triggiani M, Magaletti D, Telegrafo M, et al. An optimized feed-forward artificial neural network topology to support radiologists in breast lesions classification. InProceedings 2016 Genet Evol Comput Conf Companion. 2016. 1385-1392.
- Vaka AR, Soni B, Reddy S. Breast cancer detection by leveraging machine learning. ICT Express. 2020; 320-324.
- Abo-Seida SA. OM Keshk A Chen H. A novel deep-learning model for automatic detection and classification of breast Cancer using the transfer-learning technique. IEEE Access. 2021; 9:71194-71209.
- Ultrasound Breast Images for Breast Cancer. Kaggle.
- Breast Ultrasound Images Dataset. Kaggle.