Research Article - Onkologia i Radioterapia ( 2023) Volume 17, Issue 8
Influence of alcohol consumption and duration in the DNA repair genes polymorphisms (XRCC1 and APE1)
Asma'a H. Mohamed1, Maytham Al-Yasir2, Zainab Mahdi Al-Saygh3, Ali H. Al-Saadi4 and Mona N. Al-Terehi4*2College Of Medicine, University of Babylon, Iraq
3Faculty of Education for Girls, University of Kufa, Iraq
4College of Science, University of Babylon, Iraq
Mona N. Al-Terehi, College of Science, University of Babylon, Iraq, Email: Monanajah1981@gmail.com
Received: 09-Jul-2023, Manuscript No. OAR-23-105972; Accepted: 12-Aug-2023, Pre QC No. OAR-23-105972 (PQ); Editor assigned: 12-Jul-2023, Pre QC No. OAR-23-105972 (PQ); Reviewed: 26-Jul-2023, QC No. OAR-23-105972 (Q); Revised: 10-Aug-2023, Manuscript No. OAR-23-105972 (R); Published: 25-Aug-2023, DOI: -
Abstract
The gene- environment interaction has a major role in disease incidence and health problems, the current study aims to detect the human gene polymorphisms of X-ray Repair Cross-Complementing Group 1(XRCC1) Arg399Gln (rs25487) and Apurinic/Apyrimidinic (AP) endonuclease enzyme (APE1) (rs1130409) in alcoholism, alcohol consumption and duration in some Iraqi cases that have three levels of alcohol concentration (sub groups <50, 50-100 and >100 mg/ dl), the results show that the age and BMI were nonsignificant changes, duration and alcohol level were significant differences regarding to alcohol subgroups. The genotyping of XRCC1 showed nonsignificant association with alcoholism in compared with control group (P 0.629, 0.596), and strong association of APE1 with alcoholism (p 0.000), The alcohol level according to XRCC1 genotyping showed that AA has high level of alcohol than AG and GG in non-significant elevation (p 0.966), regarding to APE1 genotyping non-significant difference elevation in wild type than Mutated type (p 0.196) was observed. The distribution of XRCC1 genotyping according to alcoholism subgroups showed significant association (p 0.0461), and non- significant association (p 0.0614) of APE1 distribution. The XRCC1 genotyping belong to duration of alcohol consumptions showed nonsignificant association (p 0.371), and non-significant association observed in The APE1 genotyping (p 0.260). in conclusion; we can conclude that the XRCC1 genotyping didn’t associate with alcoholism and alcohol duration but significant correlated with alcohol level, while the APE1 was strong associated with alcoholism but didn’t associate with alcohol level and duration.
Keywords
XRCC1, Arg399Gln (rs25487), APE1, gene polymorphisms alcohol consumption, duration
Introduction
The DNA repair system is one of the important vital processes to genome maintenance and stability, numerous enzymes involved in the repair of DNA by different pathways, one of these enzymes is a DNA base-excision repair called human X-Ray Repair Cross-Complementing Group 1gene (XRCC1), the encoded gene located in the chromosome 19q13 [1]. the Human XRCC1 is existing in a different isoenzymes according to amino acid substitution that resulted from SNPs in XRCC1 gene namely Arg194Trp (rs1799782), Arg280His25489), and Arg399Gln (rs25487) [2], the last SNP which formed 399Gln has been shown to be associated with DNA repair capacity reduction, represent by DNA adducts persistence, that lead to different disease and health problems [3-6], on the other hand other isoenzymes Arg194Trp and Arg280His polymorphisms still under investigation. Varied activities were utilized by the base-excision repair pathway to remove non-bulky base adducts generated by oxidation, methylation and reduction by oxidative damage or ionizing radiation [7,8].
One of the base excision repair enzyme is Apurinic/Apyrimidinic (AP) endonuclease enzyme which encoded by APE1 gene, it has able to prevent transvers of base that formed by oxidized or reduced bases [9- 11] researchers reported about 18 polymorphisms in APE1 gene, notably the Asp148Glu polymorphism was well established belong to its role in the DNA repair activity alteration [12]. The rs1130409 polymorphism in APE1 is the result of T>G, T>C and T>A transition which leads to the substitution of aspartic acid, resulting in loss-of-function of APE1, DNA binding and activity of endonuclease, limitation the interaction with other base excision repair proteins and decreased oxidative damage repair [13]. The wild type of this SNP is TT and Mutated type referred to other genotyping.
Alcohol consuming has been found to be generated free radicals like Reactive Oxygen Species (ROS) causes cell component damage as well as lipid peroxidation, and acetaldehyde—that lead to DNA damage that can be repaired by the DNA base-excision repair pathway [14].
Substance abuse is Pathological use of a substance leading to significant complication demonstrated by legal, medical or psychosocial problems. Or use the substance in spite of the harmful consequences and it is one of the leading causes of death among the adolescence and young adult. Alcohol abuse is the commonest type of addiction around the world alcohol uptake produced acetaldehyde is a highly reactive has able to DNA-damaging [15]. In the Asian population the acetaldehyde detoxification is Impaired that may be associated with alcoholrelated cancers [16]. The DNA crosslink repair is used for Cells protection against acetaldehyde-induced damage, the Fanconi Anaemia (FA) is caused by impaired in this cell protection FA is a disease characterized by blood cells production failure and cancer predisposition [17,18]. However, the DNA damage nature stimulates by acetaldehyde and how this is repaired remains under investigations [19].
Current study aims to evolution the Interaction the XRCC1 Arg399Gln (rs25487) polymorphisms with alcohol consumption and duration in some Iraqi alcohol abuse.
Materials and Methods
39 cases of alcohol abuser referred by the judge because of low violation under the effect of alcohol to the forensic laboratory for estimation of alcohol level were included in this study. Alcoholism classified into three subgroups according to alcohol level, included less than 50 mg/dl, 5mg/dl,-100 mg/ dl and more than 100 mg/dl A cross sectional study was suggested for detecting two DNA repair enzymes in alcoholism in Babylon university, in the current study the XRCC1 was targeted at the SNP Arg399Gln (28152) G>A. and APE1 rs1130409, the alcoholism samples enrolled in the current study have (age ranged 19-54 years) and twenty nine healthy individuals (age range 20-40 years) as a control group, samples were taken from cases and control according to the ethical approval of ministry of environment and health in Iraq, All DNA samples were extracted by extraction kit (Favorgen), then the repair genes were detected as a following; the XRCC1 Arg399Gln (28152) G>A polymorphism was studied by CTTPPCR at annealing TM 59ºC for 30 seconds (15). F1 TCC, CTG, CGC, CGC, TGC, AGT, TTC, T; R1 TGG, CGT, GTG, AGG, CCT, TAC, CTC, C ; F2, TCG, GCG GCT, GCC, CTC, CCA; and R2 AGC, CCT, CTG, TGA, CCT, CCC, AGG C, the 447 bp of G allele (399Arg), 222 of A allele (399Gln) with 630bp common band (19) . And Belong to APE1 rs1130409 gene the primers were 5′-CCT, ACG, GCA, TAG, GTG, AGA, CC; R1:5′-TCC, TGA, TCA, TGC, TCC, TCC-3’; F2: 5′- TCT, GTT, TCA, TTT, CTA, TAG, GCG, AT; R2: 5′-GTC, AAT, TTC, TTC, ATG, TGC, CA at Tm 60ºC (19), the size of the products were (236 bp band for TT wild type and nonamplification product for Mutated type). The electrophoresis was applied with Ethidium Bromide stain utilized agarose gel 1%, 100 V, 20 mA for 40 min.
Data analysis
The results presented as mean ± SE or SD, and percentage (%), ANOVA one way, independent t test and Odd ratio (CI95%) were used for significant detection at P<0.05.
Results
The current study deal with DNA repair gene XRCC1and APE1 in chronic alcohol consumption, the alcoholism classified into three subgroups according to alcohol level, subgroups included less than 50 mg/ dl, 5mg/dl, 0-100 mg/ dl and more than 100 mg/ dl. Results showed that subgroups has 41.46, 34.14 and 24
39% respectively. Their age and BMI were non-significant changes (p 0.060, 0.405), duration and alcohol level were significant differences (p 0.041, 0.000) (Table 1).
Tab. 1. Mean differences of (age, duration, BMI, Alcohol levels and percentages) of alcoholism subgroups (mean ± SD, ANOVA one way, p less than 0.05)| Subjects | Age (year) | Duration (year) | BMI (Kg/m2) | Alcohol level (mg/ dl) | Percentage % |
|---|---|---|---|---|---|
| <50 mg/ dl | 29.00±6.70 | 3.23±1.98 | 26.23±3.05 | 47.29±4.17 | 41.46 |
| 50-100 mg/ dl | 27.78±9.90 | 3.50±3.34 | 27.76±4.21 | 63.00±6.17 | 34.14 |
| >100 mg/ dl | 36.60±11.67 | 5.90±2.60 | 26.07±3.39 | 142.00±31.43 | 24.39 |
| P value | 0.06 | 0.041 | 0.405 | 0 | - |
The genotyping of XRCC1 showed that non-significant association between XRCC1 (AG, GG and AA) (P 0.629, 0.596), and significant association of APE1 with alcoholism (p 0.000) (Table 2).
Tab. 2. the XRCC1 and APE1 genotyping distribution in alcoholism and control group (odd ratio CI 95%, P less than 0.05, RF references group)
| Genotyping | Alcoholism | Control | Odd ratio | Sig |
|---|---|---|---|---|
| XRCC1(rs25487) | ||||
| AG | 21 (52.5) | 14(48.27) | 1.2632 | 0.6293 |
| 0.4893 to 3.2612 | ||||
| GG | 14(35) | 13(44.82) | 1.5476 | 0.5968 |
| 0.3068 to 7.8067 | ||||
| AA | 5(12.5) | 3(10.34) | RG | |
| APE1 (rs1130409) | ||||
| Wild type | 87.5 | 27.28 | 18.375 | 0.0001 |
| Mutatedtype | 12.5 | 72.72 | 5.3097 to 63.5894 | |
The XRCC1 genotyping using CTTP-PCR, and APE1 genotyping (Figure 1).
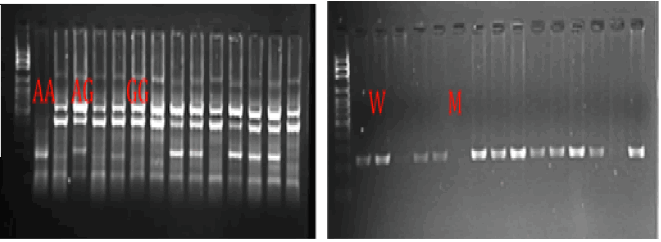
Figure 1: The XRCC1 genotyping using CTTP-PCR, and APE1 genotyping, the electrophoresis pattern at 1% agaros, 80 V, for 50 min with ethidum bromide stain, DNA ladder 100-1 kb, for XRCC1 the PCR products (GG 447 bp, AA 222 bp, AG 447+222 bp and common band 630 bp). For APE1 wild type 206 bp, Mutated type non-amplification product).
The alcohol level according to XRCC1 genotyping showed that AA has high level of alcohol than AG and GG in non-significant elevation (p 0.966) (Figure 2).
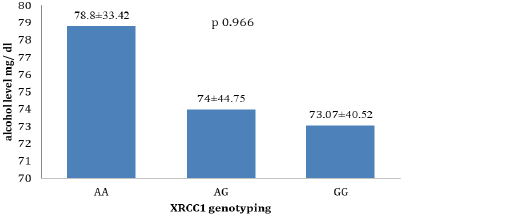
Figure 2: The alcohol level according to XRCC1 genotyping (AA, AG and GG (mean ± SD, ANOVA one way, p less than 0.05)
The alcohol level according to APE1 shows that non-significant difference elevation in wild type than Mutated type (p 0.196) (Figure 3).
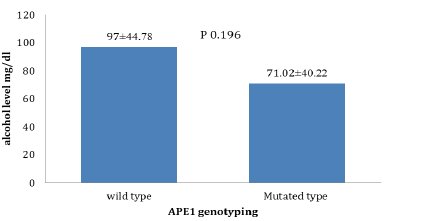
Figure 3: The alcohol level according to APE 1 genotyping (wild type and Mutated type) (mean ± SD, independent t test , p less than 0.05)
The distribution of XRCC1 genotyping according to alcoholism subgroups showed a significant association (p 0.0461) of genotyping with the level of alcohol, AG was observed in first and third subgroups (64.7%, 55.55%) respectively (Table 3).
Tab. 3. The XRCC1 genotyping distribution according to alcoholism subgroups. (X2 test, p less than 0.05)
| Alcoholism sub-group | AA% | AG% | GG% | X2 | sig |
|---|---|---|---|---|---|
| <50 mg/ dl | 5.88 | 64.7 | 29.41 | 9.683 | 0.0461 |
| 50 to 100 mg/ dl | 14,28 | 35.71 | 50 | ||
| >100 mg/ dl | 11.11 | 55.55 | 33.33 |
The distribution of APE1 genotyping belong to alcoholism subgroups showed non- significant association (p 0.0614) of genotyping with the level of alcohol (Table 4).
Tab. 4. the APE1 genotyping distribution according to alcoholism subgroups. (X2 test, p less than 0.05).
| Alcoholism sub-group | Wild type% | Mutated type% | X2 | sig |
|---|---|---|---|---|
| <50 mg/ dl | 42.85 | 40 | 5.57983 | 0.06143 |
| 50 to 100 mg/ dl | 40 | 0 | ||
| >100 mg/ dl | 17.14 | 6 |
The XRCC1 genotyping belongs to duration of alcohol consumptions shows non-significant association with duration of alcohol uptake (p 0.371) (Table 5)
Tab. 5. The XRCC1 genotyping distribution according to duration of alcohol uptake (X2 test, p less than 0.05)
| Duration | AA% | AG% | GG% | X2 | sig |
|---|---|---|---|---|---|
| <5 years | 8 | 60 | 32 | 4.26 | 0.3718 |
| 5-10 years | 16.66 | 25 | 58.33 | ||
| >10 years | 0 | 50 | 50 |
The APE1 genotyping regarding to duration of alcohol consumptions shows non-significant association with duration of alcohol uptake (p 0.260) (Table 6).
Tab. 6 The APE1 genotyping distribution according to duration of alcohol uptake (X2 test, p less than 0.05)
| Duration | Wild type% | Mutated type% | X2 | sig |
|---|---|---|---|---|
| <5 years | 68.57 | 40 | 2.69 | 0.26 |
| 5-10 years | 25.71 | 60 | ||
| >10 years | 5.71 | 0 |
Discussion
The present output exhibits that there were different factors impacted in the alcohol concentration in Blood as well as sex, BMI, the type of liqueur if alcohol used with food or drugs, like antihistamines and cimetidine (inhibits gastric alcohol dehydrogenase), phenothiazines, and metoclopramide (stimulating gastric emptying thus absorption elevation) [20]. In the current study the subgroup (<50 mg/ dl) has high percentage of samples in alcoholism, then subgroup (50-100 mg/ dl) and finally (>100 mg/ dl), on the other hand the ages of these subgroups were (29-38 years) with duration of alcohol consumption which about 3 years, this lead to thought that alcoholism is really problems among Iraqi young's cusses harmful effects and health problems, investigations clarified that the alcohol uptake for Long-term is a main risk factor of liver disease and lead to liver cirrhosis and dysfunction which stopped carcinogenic compounds detoxification [21]. Notably, in addition to ROS generating and oxidative redox disturbance, ethanol may affect the nutritional status, immune function, DNA damage, that influences the risks for various cancers [22]. The 7,8-dihydro-8- oxoguanine or 8-oxoguanine, 8-oxo-Gua is a major form of DNA oxidative damage [23], the oxidative DNA damage detection used in elucidating the cancer induction mechanisms by alcohol consumption [24, 25]. The other types of alcohol induction DNA damage is acetaldehyde that cause DNA cross link .
All forms of the DNA damages are repaired by different pathways,Hodskinson et al found two replication-coupled pathways repair of the acetaldehyde induction lesions by alcohol uptake. The first pathway operates using excision—analogous to the mechanism used to repair the interstrand crosslinks, and the second requires replication fork convergence. The current finding didn't find an association of XRCC1 with alcoholism in compared with control and this may be because the level ROS in alcoholism in some Iraqi samples didn’t change in comparison with control , also the ROS detoxification mechanism may be contributed in ROS trapping like Glutathioe S-Transferase [26]. Another study found that the XRCC1 polymorphisms have a main role in colorectal cancer risk associated with alcohol consumption [27]. Other researchers suggested that the high alcohol intake may increase colorectal cancer in the individuals have a cooperative action between the 194Trp allele or the 399Gln allele [28].
There was a significant association between XRCC1 genotyping and alcoholism subgroups (levels of alcohol), this association refer to affected XRCC1 genotyping with alcohol level, these findings agree with Yin et al and Songserm et al. [29] that found the individuals with the XRCC1 399Gln/Gln genotype have an important role in colorectal cancer risk associated with alcohol consumption. Rossit et al. agree with current finding, that significant association between the 399Gln polymorphism and the risk of liver cirrhosis in older individuals over the age of 45 years of heavy alcohol uptake [30]. In addition, significant interaction was observed between GG genotype of rs25489 polymorphism in XRCC1 and alcohol drinking to increase the risk of CRC [31]. An allele carriers of rs25487 in XRCC1 showed interaction with alcohol intake to decrease risk of Colorectal Carcinogenesis but AG genotype of rs25487 interacts with smoking to increase the Colorectal Carcinogenesis risk [32].
There were poor information about the impact of APE1 genotyping with The alcoholism, the association of APE1 with alcoholism can be considered as indirect relation, the DNA damage caused by oxidative stress that increased in alcoholism and APE1 gene targeting by Free radicals may be clarified this relation, however, Sun et al [33] found the individuals having genotype of APE1Asp148Glu TT without the habit of alcohol consumption have a 4.13 times increased risk of benzene poisoning for the alcohol user with the genotype of APE1.
The previous studies deal with gene-environment interaction in the disease incidence like alcoholism and smoking habits, the effects of alcohol uptake were varied among the population and its depended on several factors like nutrition, genetic predisposition and gene –environment interaction [34-38].
Conclusion
The current findings concluded that the no-association of the XRCC1 Arg399Gln (rs25487) genotyping with alcoholism, didn’t confirm of XRCC1 with alcohol levels in drunks, strong association with APE 1 (rs1130409) genotyping with alcoholism and didn't associate with duration and alcohol level. However, we need more investigations about other SNPs relations.
References
- Mohrenweiser HW, Carrano AV, Fertitta A, Perry B, Thompson LH, et al. Refined mapping of the three DNA repair genes, ERCC1, ERCC2, and XRCC1, on human chromosome 19. Cytogenet Cell Genet. 1989;52:11–14.
[Google Scholar] [ CrossRef]
- Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–608.
[Google Scholar] [ CrossRef]
- WarchoÅ? T, Mostowska A, Lianeri M, ÅÄ?cki JK, JagodziÅ?ski PP. XRCC1 Arg399Gln gene polymorphism and the risk of systemic lupus erythematosus in the Polish population. DNA and cell biology. 2012;31:50-56.
- Lunn RM, Langlois RG, Hsieh LL, Thompson CL, Bell DA. XRCC1 polymorphisms: effects on aflatoxin B1-DNA adducts and glycophorin A variant frequency. Cancer Res.1999;59:2557–2561.
[Google Scholar] [ CrossRef]
- Duell EJ, Wiencke JK, Cheng TJ, Varkonyi A, Zuo ZF, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cells. Carcinogenesis. 2000;21:965–971.
- Yin G, Morita M, Ohnaka K, Toyomura K, Hamajima N,et al. Genetic polymorphisms of XRCC1, alcohol consumption, and the risk of colorectal cancer in Japan. J Epidemiol. 2012;22:64-71. Google Scholar CrossRef
- Beckman KB, Ames BN. Oxidative decay of DNA. J Biol Chem. 1997;272:19633–19636.
[Google Scholar ] [CrossRef]
- Ladiges W, Wiley J, MacAuley A. Polymorphisms in the DNA repair gene XRCC1 and age-related disease. Mech Ageing Dev. 2003;124:27–32.
- Ito H, Matsuo K, Hamajima N, Mitsudomi T, Sugiura T, et al. Gene–environment interactions between the smoking habit and polymorphisms in the DNA repair genes, APE1 Asp148Glu and XRCC1 Arg399Gln, in Japanese lung cancer risk. Carcinogenesis. 2004;25:1395-401.
[Google Scholar] [ CrossRef]
- Lu L, Zhu C, Xia B, Yi C. Oxidative demethylation of DNA and RNA mediated by non-heme iron-dependent dioxygenases. Chem Asian J.2014;9:2018-2029.
[Google Scholar] [ CrossRef]
- Sliwinski T, Przybylowska K, Markiewicz L, Rusin P, Pietruszewska W et al. MUTYH Tyr165Cys, OGG1 Ser326Cys and XPD Lys751Gln polymorphisms and head neck cancer susceptibility: a case control study. Mol Biol Rep. 2011;38:1251-1261.
[Google Scholar] [ CrossRef]
- Zhong JH, Zhao Z, Liu J, Yu HL, Zhou JY, et al.Association between APE1 Asp148Glu polymorphism and the risk of urinary cancers: a meta-analysis of 18 case-control studies. OncoTargets Ther. 2016;9:1499-510.[ Google Scholar] [ CrossRef]
- Hadi MZ, Coleman MA, Fidelis K, Mohrenweiser HW, Wilson DM 3rd. Functional characterization of APE1 variants identified in the human population. Nucleic Acids Res. 2000;28:3871-9.
[Google Scholar] [ CrossRef]
- Rossit AR, Cabral IR, Hackel C, da Silva R, Froes ND, et al. Polymorphisms in the DNA repair gene XRCC1 and susceptibility to alcoholic liver cirrhosis in older Southeastern Brazilians. Cancer Lett. 2002;180:173–182.[ Google Scholar] [ CrossRef]
- Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6. Google Scholar CrossRef
- Lai CL, Yao CT, Chau GY, Yang LF, Kuo TY,et al. Dominance of the inactive Asian variant over activity and protein contents of mitochondrial aldehyde dehydrogenase 2 in human liver. Alcohol Clin Exp Res. 2014;38:44–50.
[Google Scholar] [ CrossRef]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53-58.
[Google Scholar] [ Crossref]
- Garaycoechea JI, Patel KJ. Why does the bone marrow fail in Fanconi anemia? Blood. 2014;123:26-34.
[Google Scholar] [ Crossref]
- Hodskinson MR, Bolner A, Sato K, et al. Alcohol-derived DNA crosslinks are repaired by two distinct mechanisms.Nature.2020;579:603-608.
[Google Scholar] [ Crossref]
- Ito H, Matsuo K, Hamajima N, Mitsudomi T, Sugiura T, et al. Gene-environment interactions between the smoking habit and polymorphisms in the DNA repair genes, APE1 Asp148Glu and XRCC1 Arg399Gln, in Japanese lung cancer risk. Carcinogenesis. 2004;25:1395-1401.
[Google Scholar] [ Crossref]
- Paton A. Alcohol in the body. BMJ (Clinical research ed.). 2005;330:85-87.
[Google Scholar] [ Crossref]
- Osna NA, Donohue TM Jr, Kharbanda KK. Alcoholic Liver Disease: Pathogenesis and Current Management. Alcohol res.: curr. rev.2017;38:147-161.
[Google Scholar] [ Crossref]
- Hirano T. Alcohol consumption and oxidative DNA damage. International journal of environmental research and public health.2011;8:2895-2906.
[Google Scholar] [ Crossref]
- Kasai H, Nishimura S. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res. 1984;12:2137-2145.
[Google Scholar] [ Crossref]
- Hirano H, Hirano T, Hirata T, Tamura M, Yamaura T, et al. Experimental liver fibrosis induced in rats receiving high doses of alcohol and alternating between regular and vitamin-depleted diets. Experientia. 1996;52:710-715.
[Google Scholar ] [Crossref]
- Alkadir OKA, Al-Mashhadani ZI, Al-Terehi MN, Al-Rrubaei HA, Alkaim AF. The Estimation of Oxidative Stress from Alcohol Use Disorders in Iraqi Population. International Journal of Pharmaceutical Quality Assurance. 2021;12:300-302.
[Google Scholar ] [Crossref]
- Al-Terehi MN, Altimari US, Kadhim AJ, Kadhum AM. The Glutathione S-Transferasee (GSTT and GSTM) Genotyping and Alcohol Level in Drunks. Int. J. Pharm. Qual. Assur.,2021;12(4):331-333.
[Google Scholar] [ Crossref]
- Yin G, Morita M, Ohnaka K, Toyomura K, Hamajima N, et al. Genetic polymorphisms of XRCC1, alcohol consumption, and the risk of colorectal cancer in Japan. J. epidemiol. 2012;22:64-71.
[Google Scholar] [ Crossref]
- Gao CM, Ding JH, Li SP, Liu YT, Cao HX, et al. . Polymorphisms in XRCC1 gene, alcohol drinking, and risk of colorectal cancer: a case-control study in Jiangsu Province of China. Asian Pac. j. cancer prev.. 2013;14:6613-6618.
[Google Scholar] [ Crossref]
- Songserm N, Promthet S, Pientong C, Ekalaksananan T, Chopjitt P, et al.Gene–environment interaction involved in cholangiocarcinoma in the Thai population: polymorphisms of DNA repair genes, smoking and use of alcohol. BMJ open. 2014;4.
[Google Scholar] [ Crossref]
- Rossit AR, Cabral IR, Hackel C, da Silva R, Froes ND, et al.Polymorphisms in the DNA repair gene XRCC1 and susceptibility to alcoholic liver cirrhosis in older Southeastern Brazilians. Cancer Lett. 2002;180:173-182.
[Google Scholar] [ Crossref]
- Yin G, Morita M, Ohnaka K, Toyomura K, Hamajima N, et al.Genetic polymorphisms of XRCC1, alcohol consumption, and the risk of colorectal cancer in Japan. J Epidemiol. 2012;22:64-71.
[Google Scholar] [ Crossref]
- Liu J, Zheng B, Li Y, Yuan Y, Xing C. Genetic Polymorphisms of DNA Repair Pathways in Sporadic Colorectal Carcinogenesis. J Cancer.2019;10:1417-1433.
[Google Scholar] [ Crossref]
- Sun P, Zhang ZB, Wan JX, Jin XP, Xia ZL. Relationship of genetic polymorphism in APE1 and ADPRT to risks of chronic benzene poisoning. 2006;24(7):385-389.
[Google Scholar ] [Crossref]
- Barr PB, Salvatore JE, Maes H, Aliev F, Latvala A, et al. Education and alcohol use: A study of gene-environment interaction in young adulthood. Soc Sci Med. 2016;162:158-167.
[Google Scholar] [ Crossref]
- Latvala A, Dick DM, Tuulio-Henriksson A, Suvisaari J, Viken RJ,et al. Genetic correlation and gene–environment interaction between alcohol problems and educational level in young adulthood. J stud alcohol drugs,2011;72:210-220.
[Google Scholar] [ Crossref]
- Al-Terehi MN, Zahraa Haleem Alqaim, Arafat Hussein Aldujaili. Impact of DNA Repair System Genes RAD-18 and XRCC1 Polymorphism in Depression Disorders Patients. Clin Schizophr Relat Psychoses. 2021;15.
[Google Scholar] [ Crossref]
- Al-Terehi MN, Altimari US, Kadhim AJ, Al-Rrubaei HA. The Combination Between Anti-depressant and Anti-diabetic Therapy Effects in Depressed Patients with Type 2 Diabetic Mellitus. International Journal of Pharmaceutical Quality Assurance. 2021;12:287-289.
[Google Scholar] [ Crossref]