Research Article - Onkologia i Radioterapia ( 2023) Volume 17, Issue 3
Monoclonal pattern of T-Lymphocytes and B-Lymphocytes receptors gene rearrangement in Acute Lymphocytic Leukemia in Basrah
Nibras S. Al-Ammar1*, Hermann Kreyenberg2 and Mayada T. Abdulrahman12MRD-Chimarism Laboratory, Stem Cell Transplantation Centre, Goethe University Hospital, Frankfurt, Germany
Nibras S. Al-Ammar, Microbiology Department, College of Medicine, University of Basrah, Basrah, Iraq, Email: nibrasalammar@yahoo.com, alammar.nibras@gmail.com
Received: 08-Mar-2023, Manuscript No. OAR-23-91142; Accepted: 27-Mar-2023, Pre QC No. OAR-23-91142 (PQ); Editor assigned: 10-Mar-2023, Pre QC No. OAR-23-91142 (PQ); Reviewed: 24-Mar-2023, QC No. OAR-23-91142 (Q); Revised: 26-Mar-2023, Manuscript No. OAR-23-91142 (R); Published: 28-Mar-2023
Abstract
Paediatrics clinicians face a problem in evaluation of chemotherapy treatment used for Acute Lymphoblastic Leukaemia (ALL) patients in Iraq. They need an accurate test to evaluate the treatment. This study aimed to detect the clonality pattern of T and B lymphocytes receptors gene rearrangements in ALL patients in Basrah before chemotherapy. Bone morrow aspiration samples from 50 ALL patients used for DNA extraction followed by multiplex PCR by using 42 primers. A total of 147 bands cut from polyacrylamide gel and sent for sequencing in order to detect the pattern of receptors gene rearrangements. 15th ALL patients were found to have rearrangements in the studied genes coding for receptors of T and B lymphocytes; TCR and Ig. Ten patients have a rearrangement of Ig while 5 patients rearranged TCR. Immunoglobulin Heavy Chain (IGH) gene was rearranged in 6 patients. Some patients appeared to have more than one rearrangement in TCR, IGH, IGK, TCRD and TCRG genes, while the rest of the samples have only one rearrangement. The highest clonality appeared in IGH gene in VH1-7/JH, DH1-6/JH and DH7/JH. Studying Ig and TCR gene rearrangements are an important step for testing MRD which consider now a diagnostic method that evaluate chemotherapy protocol treatment and also predict the patient chance of relapse. The current study might be the first step for further work that can help in chemotherapy treatment evaluation in Iraq.
Keywords
monoclonal, TCR, IG, gene rearrangement, multiplex PCR, acrylamide gel, ALL
Introduction
Leukaemia is characterized by the growth and differentiation of leukocytes abnormally [1, 2]. Acute lymphoblastic leukaemia considered one of serious paediatric malignancies [3]. T-cell Receptors (TCRs) were first described based on their resemblance to immunoglobulin DNA sequences. These are heterodimeric polypeptide chains having constant and variable portions [4]. Acute lymphoblastic leukaemia treatment has made great progress, as have new molecular techniques for monitoring ALL patients throughout their disease genes. It has IGK and TCR genes that rearranged during their development and used to determine clonality and minimum residual illness [5]. Clonality testing substantially aids and facilitates the detection of lymphoid cancers. The T/B cell receptor rearrangement testing based on multiplex PCR is now utilized as a clinical technique to diagnose probable lymphoproliferative illness [6]. Clonal Variable Diversity Joining (VDJ) rearrangement that happens through mutation was used as a marker to indicate the clonality of B and T cells during their maturation [7]. The junctional regions are thought to represent distinct fingerprint-like sequences [8]. Clonal Ig and TCR rearrangements has found practical use of Minimal Residual Disease (MRD) detection, particularly in ALL [9], also have an important role in identifying targets for immunotherapy [10]. The current study aimed to study monoclonal pattern of T and B lymphocytes receptors gene rearrangements in ALL patients in Basrah before chemotherapy. Detection of Ig and TCR clonality patterns is recommended in ALL patients in Basrah. That will make it possible to obtain a reference value in MRD testing.
Methods
A cohort study carried out from 2020 to 2022 and involved 50 ALL children in Basrah Children Hospital/oncology department, their ages ranged from 1 month-15 years. The inclusion criteria were newly diagnosed children with ALL according to clinical background of each patient includes parameters such as HB, WBC, platelets, blood film and flow cytometry. The exclusion criteria were children with other types of haematological malignancy. Ethical consideration of the approval of the Basrah health authority was obtained previously
Fresh bone morrow samples (2 ml-3 ml) were collected in a sterile plastic tubes, from newly diagnosed ALL patients which was already available for routine diagnosis in the haematology laboratory of Basrah Children Specialty Hospital. Following the manufacturer's instructions, Specific kits for extraction and purification of DNA were used (Wizard Genomic DNA Purification Kit/Promega Corporation/USA). Measuring DNA concentration purity was done by Quantiflour dsDNA system/ Promega/USA by Quantus Flourometer. Diluting the DNA Dye 1:400 in 1X TE buffer, then mixed the dye 0.5 µl with 200 µl of the buffer. The blank was prepared by adding 200µl of working solution into a 0.5 ml PCR tube (kept in a dark place). The standard calibration was prepared by adding 10 µl of diluted standard to 200 µl of DNA dye working solution in a 0.5 ml tube, mixed and kept away from light. Then 2 µl of the sample was added to 200 µl of (DNA dye working solution) in a 0.5 ml tube and was vortexed well and incubated for 5 minutes in dark place. Fluorescence of the sample was measured and recorded the final sample concentration result. The DNA integrity was investigated by gel electrophoresis.
Multiplex Pcr Amplification
Primers ordered from Alpha DNA Company/Canada (Tables 1)A stock of several primers was prepared by taking 10 µl or 5µl of each one. The protocol for primer mixing depends on the number of primers that were used. All the 7 forward primers from stock were combined, 5 µl was taken (35 µl primer mix) 65 µl TE-buffer was added so the final primer mix was 100 µl each primer 5 pmol/ µl. For the reverse primers all the 5 primers were combined the result was 25 µl then 75 µl of TE-buffer was added to obtain a 100 µl primer mix, each primer has a concentration of 5 pmol/ µl. After the forward and reverse primers were prepared 1:1 mix from them and 2µl was added to the 25 µl PCR reaction.
Tab. 1. The sequences of primers
| Family | Gene | primer | Sequence |
|---|---|---|---|
| VH | IGH | VH1 | 5’TGGAGCTGAGCAGCCTGAGATCTGA3’ |
| VH2 | 5’CAATGACCAACATGGACCCTGTGGA3’ | ||
| VH3 | 5’TCTGCAAATGAACAGCCTGAGAGCC3’ | ||
| VH4 | 5’ GAGCTCTGTGACCGCCGCGGACACG 3’ | ||
| VH5 | 5’ CAGCACCGCCTACCTGCAGTGGAGC 3’ | ||
| VH6 | 5’ GTTCTCCCTGCAGCTGAACTCTGTG 3’ | ||
| VH7 | 5’ CAGCACGGCATATCTGCAGATCAG 3’ | ||
| JH | IGH | JH | 3’ CCAGTGGCAGAGGAGTCCATTC 3’ |
| DH | IGH | DH1 | 5’ GGCGGAATGTGTGCAGGC 3’ |
| DH2 | 5’ GCACTGGGCTCAGAGTCCTCT 3’ | ||
| DH3 | 5’ GTGGCCCTGGGAATATAAAA 3 | ||
| DH4 | 5’ AGATCCCCAGGACGCAGCA 3’ | ||
| DH5 | 5’ CAGGGGGACACTGTGCATGT 3’ | ||
| DH6 | 5’ TGACCCCAGCAAGGGAAGG 3’ | ||
| DH | IGH | DH7 | 5’ CACAGGCCCCCTACCAGC 3’ |
| VK | IGK | Vk 1f/6 | 5’ TCAAGGTTCAGCGGCAGTGGATCTG 3’ |
| VK2f | 5’ GGCCTCCATCTCCTGCAGGTCTAGTC 3’ | ||
| VK3f | 5’CCCAGGCTCCTCATCTATGATGCATCC 3’ | ||
| VK4f | 5’CAACTGCAAGTCCAGCCAGAGTGTTTT3’ | ||
| VK5 | 5’CCTGCAAAGCCAGCCAAGACATTGAT3’ | ||
| VK7 | 5’GACCGATTTCACCCTCACAATTAATCC 3’ | ||
| JK | IGK | Jk 1-4 | 3´ CCCTGGTTCCACCTCTAGTTTGCATTC 5’ |
| JK 5 | 3’CCCTGTGCTGACCTCTAATTTGCATTC 5’ | ||
| Kde | IGK | Kde | 3’ATCCTGTTGGACGAGACTGGAGACTCC5’ |
| Vɣ | TCRG | Vɣ I f | 5’GGAAGGCCCCACAGCRTCTT 3’ |
| Vɣ 10 | 5’ AGCATGGGTAAGACAAGCAA 3’ | ||
| Vɣ9 | 5’ CGGCACTGTCAGAAAGGAATC 3’ | ||
| Vɣ11 | 5’CTTCCACTTCCACTTTGAAA 3’ | ||
| Jɣ | TCRG | Jɣ1.1/2.1 | 3’CGAGTATCATTGAAGCGGACCATT 5’ |
| Jɣ1.3/2.3 | 3’ GAGAAACCGTCACCTTGTTGTG 5’ | ||
| Vδ | TCRD | Vδ1 | 5’ ATGCAAAAAGTGGTCGCTATT 3’ |
| Vδ2 | 5’ ATACCGAGAAAAGGACATCTATG 3’ | ||
| Vδ3 | 5’ GTACCGGATAAGGCCAGATTA 3’ | ||
| Vδ4 | 5’ ATGACCAGCAAAATGCAACAG 3’ | ||
| Vδ5 | 5’ ACCCTGCTGAAGGTCCTACAT 3’ | ||
| Vδ6 | 5’ CCCTGCATTATTGATAGCCAT 3’ | ||
| Jδ | TCRG | Jδ1 | 3’ CTTGGGCACACTGACACCTTG 5’ |
| Jδ2 | 3’ CTTGTGTTGAGTAGCACCTTG 5’ | ||
| Jδ3 | 3’ GAGAAGCACCTCGGGGCACTC 5’ | ||
| Jδ4 | 3’CCTTGGATAGACCTCCATGTT 5’ | ||
| Dδ2 | 5’ AGCGGGTGGTGATGGCAAAGT 3’ | ||
| Dδ3 | 3’ TATAGGAGTGGGACCCAGGGT 5’ |
Multiplex PCR had done to 50 patients according to the protocol BIOMED-2 [11]. The 42 primers were multiplexed in ten reactions for testing IGH, IGK, TCRD, TCRG rearrangements. Master Mix proportions for multiplex PCR were as followed; 2 µl Primer, 12.5 µl master mix, 10 µl H2O, 5 µl DNA and the final volume was 25 µl. In multiplex PCR protocol the temperature for pre-activation was 95°C (7 minutes), a step of denaturation at the same temperature (45 seconds), annealing step at a temperature exceed to 60°C (45 seconds). The temperature then changed to 70°C for extension step (1.30 minutes) for 35 cycles. The final extension at 70°C for 15 min., then hold at 15°C for 4 min-10 min. Electrophoresis by polyacrylamide gel (8%) was done for the PCR products. After completion of the electrophoresis the gel is subjected to staining to visualize PCR band patterns. Then DNA bands were removed from the gel by agarose band cutter and purification by centrifugation was done following the kit instructions steps and then DNA stored at 4°C or -20°C. The DNA purified from 147 bands/ 50 patients, 42 pr imers (multiplex PCR) was sent to Macrogen Company in South Korea for sequencing The sequencing was at both directions with the same oligonucleotides used in multiplex PCR reaction. Analysis of sequencing results had done by online using NCBI Ig BLAST (Figure 1 A,B).
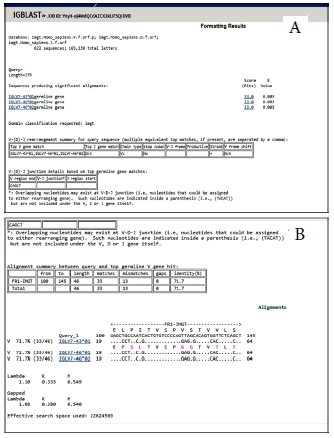
Figure 1: A,B. Result of analysis of the sequences online by using Ig Blast
Results
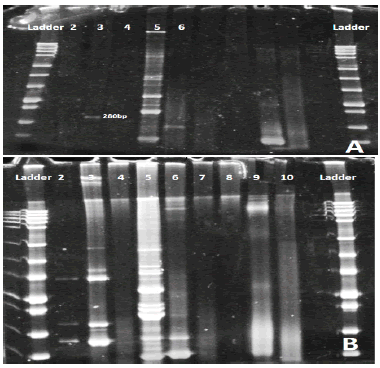
Concentration purity of genomic DNA by Quantiflour dsDNA system, varied between (3 ng/µl) and (80 ng/µl). As a result of ten multiplex PCR reactions and electrophoresis by polyacrylamide gel (8%) for detecting IGH, IGK, TCRD and TCRG rearrangements, different genes appeared in each patient but DH7 appeared in all of them because it considered as a positive control (Figure 2A and Figure 2B).
Figure 2: Polyacrylamide gel: Lane 2 DH1-6 /JH primers were used and the gel appeared negative. Lane3 VH1-7/JH primers were used and one band appear with about 280bp. Lane4 DH7/JH primer was used and appeared negative. Lane 5 VK1-7/kde were used about 6 bands appear positive for this gene. Lane 6 VK1-7/JK primers were used. 2B. Polyacrylamide gel: Lane 2 DH1-6/JH negative. Lane3 positive for VH1-7. Lane4 negative for DH7. Lane5 positive for VK1-6/kde. Lane6 positive for VK1-6/JK. Lane7 positive. Lane8, 9,10 are negative.
No rearrangement, B. Rearrangement appeared in VK1-7/Kde, C. Rearrangement in (Vδ2/Dδ3), D. Rearrangement appeared in DH3/JH, E. Rearrangement appeared in Dδ2/Dδ3, F. Rearrangement in (Vγ9 Vγ11/Jγ)
Out of 50 patients, 147 bands had sent to sequencing. 15th patients have rearranged Ig and TCR gene are while most of the samples have germline (Figure 3A). Majority of the patients have a rearrangement of immunoglobulins while the rest of the 5 patients have rearrangement of T cell receptors. Out of 15 patients, 6 cases had clonal IGH gene rearrangement in VH1-7/ JH, DH1-6/JH, DH7/JH, 4 patients have a rearrangement of clonal IGK gene in VK1-7/JK, VK/Kde (Figure 3B), 3 had delta T cell rearrangement TCRD in VγIf, Vγ10, Vγ9, Vγ11 and only 2 had gamma T cell receptor rearrangement TCRG in Vδ1-6/ Jδ1-4, Dδ2-Dδ3 and Vδ2/Dδ3 (Figure 3C). Some patients showed more than one rearrangement in both IGH (DH3/JH) (Figure 3D) and TCR genes (Dδ2/Dδ3) (Figure 3E) rearrangement (biclonal). Also there was rearrangements in both IGH (DH1-6/JH), (VH1-7/ JH), and IGK (VK1-7/JH) genes, rearrangement appeared in TCRD (Vδ2-Dδ3) and TCRG (Vγ9, Vδ11/ Jɣ1.1/2.1, Jɣ1.3/2.3) genes (Figure 3F), while the rest of the samples have only one rearrangement (monoclonal). The DH7- JH primers had mixed separately in a different tube the expected size product was 100 bp-130 bp. This primer used as a positive control in Ig and TCR rearrangements so it appeared in all patients.
Figure 3: No rearrangement, B. Rearrangement appeared in VK1-7/Kde, C. Rearrangement in (Vδ2/Dδ3), D. Rearrangement appeared in DH3/JH, E. Rearrangement appeared in Dδ2/Dδ3, F. Rearrangement in (Vγ9 Vγ11/Jγ)
Discussion
Study the monoclonal pattern of T lymphocytes and B lymphocytes receptors gene rearrangement among all children has not performed yet in Iraq. It is considered an important tool for MRD/PCR [11]. Sequencing provides a clear understanding for what happens at the level of genes during the course of the malignancies [12]. The disease results in many symptoms and many infections as a result of abnormal bone marrow functions [13], with increased blast cell production [14]. Sequencing analysis is the most dependable way to decide the development or remain of Ig and TCR rearrangement during the sickness phase [12]. Rearrangements of VDJ gene segments create massive clones, the insertion and deletion of nucleotides increases the diversity of clones, and the TCR and Ig gene rearrangement patterns of the same clone were revealed and utilized to identify a specific clone [5]. Each lymphocyte has a unique changeable antigen-receptor gene so clonality testing is relevant to all lymph proliferations. The sequential development makes the segments (V-, D-, and J) join together from numerous accessible segments, resulting in a wide diversity of antigen receptors [15]. It is important to note that mono-clonality does not always suggest malignancy. There is no published information on the Ig and TCR genes clonality style in Iraqi children with Acute Lymphoblastic Leukaemia. There were 46 B-precursor ALL patients (92%) and four T-ALL patients among the ALL patients, indicating that B-ALL was more common. Clonal pattern was found in most cases (40%) which were in agreement with other studies [16, 17]. In this study we observed IGK gene clonal pattern in 27% of the patients in VK1- 7/JK, VK/Kde. The incidence of TCRG rearrangement were low in B-ALL patients 13% using specific primers VγIf, Vγ10, Vγ9, Vγ1, our findings were equivalent to those of previous Brazilian investigations [18]. Many reasons lead to differences in clonality between nations, including the use of various methods, variations in socioeconomic position, ethnic origin, and continual immune system activation owing to viral and bacterial illnesses in Iraqi youngsters compared to those in industrialized nations. Regarding to the bands appeared after gel electrophoresis (either horizontal or vertical), not every band supposed to be a rearrangement, most bands refer to the incomplete IGH screen (DH7-JH1). This helps as a control of the PCR. As many bands appeared on the gel, and in order to reduce sequencing, it was better to run DH7-JHcons on a separate lane in order to show patterns of the germline which helps in identification of possible rearrangement. Sometimes banding results in complex structure due to the heterogeneous nature of some leukaemia. It appeared as several clones at the same line. The step followed gel electrophoresis in which bands cut and sent to sequencing were very critical. Suitable steps through multiplex PCR, gel electrophoresis, cutting the bands, purification, and sequencing showed good results in many samples. Regarding to the primers used in sequencing, these primers were the same primers that used in PCR in order to detect the exact rearrangement. Interpretation of Sequence have done online by checking the sequence by using Blast to get information about locus and if rearrangement is existing already then check with Ig-Blast to do interpretation of every sequence as a junction. Some sequences at 5’ and 3’ did not aligned so it was important to check if the rearrangement already exists by checking Blast first. Some samples showed nice bands but sequencing results gave a picture of overlay of several sequences. That was because of failing in hetero-duplex analyses, as functional rearrangements of the background with the same sizes and run on the gel together. The characteristic features of (ALL) are the blasts as proliferated immature hematopoietic cells that present in the bone marrow of patient.
Conclusion
The majority of patients have rearrangement of immunoglobulins, the highest clonality appeared in IGH gene in VH1-7/JH, DH1- 6/JH and DH7/JH. Studying Ig and TCR gene rearrangements are an important step for testing MRD which consider now a diagnostic method that evaluate chemotherapy protocol treatment and also predict the patient chance of relapse. Clonality detection is needed to solve the problem of relapse in children with leukaemia to insure that the patient is in complete remission before continued chemotherapy. The current study might be the first step for further work that can help in chemotherapy treatment evaluation in Iraq.
Acknowledgment
The researchers appreciate the time and effort invested in delivering valuable training program in (Chimerism laboratory/ Stem Cell Transplantation centre/ Goethe University Hospital/ Frankfurt/ Germany) for the molecular work and analysis of the results related to the current study
References
- Chudova DI, Sehnert AJ, Bianchi DW. Copy-number variation and false positive prenatal screening results. N Engl J Med. 2016;375:97-98.
- Bispo JA, Pinheiro PS, Kobetz EK. Epidemiology and etiology of leukaemia and lymphoma. Cold Spring Harb Perspect Med. 2020;10: a034819.
- Montaño A, Forero-Castro M, Marchena-Mendoza D, Benito R, Hernández-Rivas JM. New challenges in targeting signaling pathways in acute lymphoblastic leukaemia by NGS approaches: An update. Cancers. 2018; 10:110.
- Clambey ET, Davenport B, Kappler JW, Marrack P, Homann D. Molecules in medicine mini review: the αβ T cell receptor. J Mol Med. 2014 Jul; 92:735-41.
- Poopak B, Saki N, Purfatholah AA, Najmabadi H, Mortazavi Y, et al. Pattern of immunoglobulin and T-cell receptor-δ/γ gene rearrangements in Iranian children with B-precursor acute lymphoblastic leukaemia . Hematology. 2014; 19:259-266.
- Langerak AW, Groenen PJ, Brüggemann M, Beldjord K, Bellan C, et al. EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leukaemia . 2012; 26:2159-2171.
- Watson CT, Steinberg KM, Huddleston J, Warren RL, Malig M, et al. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet. 2013; 92:530-546.
- Van der Velden VH, Hochhaus A, Cazzaniga G, Szczepanski T, Gabert J, et al. Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects. Leukaemia . 2003; 17:1013-1034.
- Brüggemann M, Raff T, Kneba M. Has MRD monitoring superseded other prognostic factors in adult ALL?. Blood J Am Soc Hematol. 2012; 120:4470-4481.
- Klinger M, Benjamin J, Kischel R, Stienen S, Zugmaier G. Harnessing T cells to fight cancer with BiTE® antibody constructs–past developments and future directions. Immunol. Rev. 2016; 27:193-208.
- Van Dongen JJ, Langerak AW, Brüggemann M, Evans PA, Hummel M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukaemia . 2003; 17:2257-2317.
- Kreyenberg H, Eckert C, Yarkin Y, Reising M, Willasch A, et al. Immunoglobulin and T-cell receptor gene rearrangements as PCR-based targets are stable markers for monitoring minimal residual disease in acute lymphoblastic leukaemia after stem cell transplantation. Leukaemia . 2009; 23:1355-1358.
- Hastings CA, Torkildson JC, Agrawal AK. Evaluation of the Child with a Suspected Malignancy. Pediatric Clinics of North America. 1991; 38:223-248.
- Shahab F, Raziq F. Clinical presentations of acute leukaemia . J Coll Physicians Surg Pak. 2014 Jul 1;24:472-476.
- Van Krieken JH, Langerak AW, Macintyre EA, Kneba M, Hodges E, et al. Improved reliability of lymphoma diagnostics via PCR-based clonality testing:—Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukaemia . 2007; 21:201-206.
- Szczepanski T, Willemse MJ, Brinkhof B, van Wering ER, van der Burg M, et al. Comparative analysis of Ig and TCR gene rearrangements at diagnosis and at relapse of childhood precursor-B–ALL provides improved strategies for selection of stable PCR targets for monitoring of minimal residual disease. Blood J Am Soc Hematol. 2002; 99:2315-2323.
- Dawidowska M, Derwich K, SzczepaÅ?ski T, JóÅ?kowska J, Van der Velden VH, et al. Pattern of immunoglobulin and T-cell receptor (Ig/TCR) gene rearrangements in Polish pediatric acute lymphoblastic leukaemia patients—implications for RQ-PCR-based assessment of minimal residual disease. Leuk Res. 2006;30:1119-1125.
- Scrideli CA, Kashima S, Cipolloti R, Defavery R, Tone LG. Clonal evolution as the limiting factor in the detection of minimal residual disease by polymerase chain reaction in children in Brazil with acute lymphoid leukaemia . J Pediatr Hematol/Oncol. 2002; 24:364-367.